

The 1000 Genomes Project Long-read Sequencing Consortium (1KGP-LRSC) is building on the landmark work done by the 1000 Genomes Project (1KGP), which began in 2008 as a collaborative initiative to establish a database of normal human genetic variation by sequencing the genomes of over a thousand healthy individuals from diverse ancestries. In the end, the 1KGP study sequenced over 3,000 genomes using short-read sequencing, and it continues to provide invaluable insights into human genetic diversity.
The 1KGP-LRSC kicked off on Thursday, June 30, 2022. We aim to perform long-read sequencing of all 3,202 1KGP samples, which are available as DNA or cell lines from the NHGRI's Sample Repository for Human Genetic Research housed at the Coriell Institute for Medical Research. To obtain high-coverage, high-quality long-read assemblies, we are isolating high molecular weight DNA directly from cell culture of 1KGP cell lines obtained from Coriell.
The goal of the Consortium is to identify a broader spectrum of genomic variation than is possible using short-read sequencing so we may further improve our understanding of human genetic disease. This dataset is already enabling us to better understand normal patterns of human structural variation, identify variation in difficult-to-map regions of the genome, and study repeat expansions and methylation patterns.

We are a collaborative group of researchers from around the world interested in leveraging long-read sequencing to better understand the normal patterns of structural variation, methylation, and repeat expansion in the population so we can more effectively identify missing disease-causing variation in individuals. The project is led by Danny Miller and Evan Eichler at the University of Washington, and cell culture and DNA extraction are performed in the Miller and Eichler labs. Sequencing is performed at the University of Washington, the New York Genome Center, and Stanford University. Individuals and institutions contributing to this work are listed below. If you would like to be included here, please let us know.

Long-read sequencing is being performed using both Oxford Nanopore and PacBio. Nanopore technology detects changes in current as single-stranded DNA or RNA molecules pass through a protein pore. The first 100 samples were sequenced on the R9.4.1 pore, and subsequent samples were sequenced with the higher accuracy R10 chemistry. PacBio sequencing identifies DNA bases through real-time detection of fluorescently labeled nucleotides. Both platforms support direct detection of epigenetic modifications, including DNA methylation. Data from both platforms will be integrated through a joint analysis pipeline that harmonizes variant calls, resolves discrepancies, and consolidates epigenetic signatures to produce a comprehensive genomic profile.

The 1KGP-LRSC is committed to publicly releasing data as they are generated, after basecalling and standard QC. Raw Nanopore sequencing data, processed data, and summary data can be found on AWS.
Analysis of the first 100 genomes sequenced for this project on the Nanopore platform is published in Genome Research (PMCID PMC11610458).
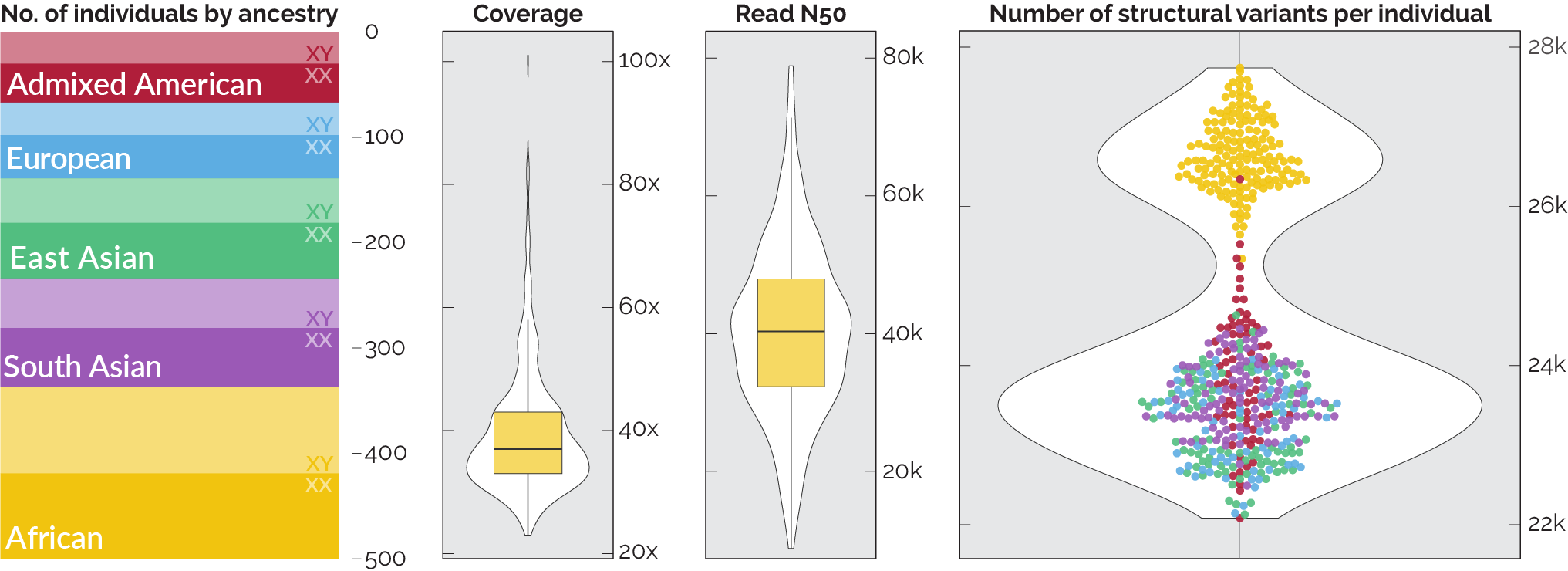
We have since completed sequencing and basic analysis of 500 samples and are beginning to generate PacBio data as well, which will be released in batches as it is available. The 500 Nanopore-sequenced samples:

The 1KGP-LRSC is open to all. Please contact Danny Miller if you are interested in joining the consortium and to be added to the Slack group.