

The University of Washington Nanopore Sequencing Core provides DNA and RNA sequencing services on the Oxford Nanopore Technologies platform. For collaborators, we perform library preparation, sequencing, and analysis from a variety of sample inputs, including blood, saliva, buccal swabs, and cell pellets. We also accept direct DNA or RNA submissions.
This page provides detailed information on available services and how to prepare, package, and ship your samples. Before submitting samples, we ask that you contact us for a consultation and to receive an estimate.
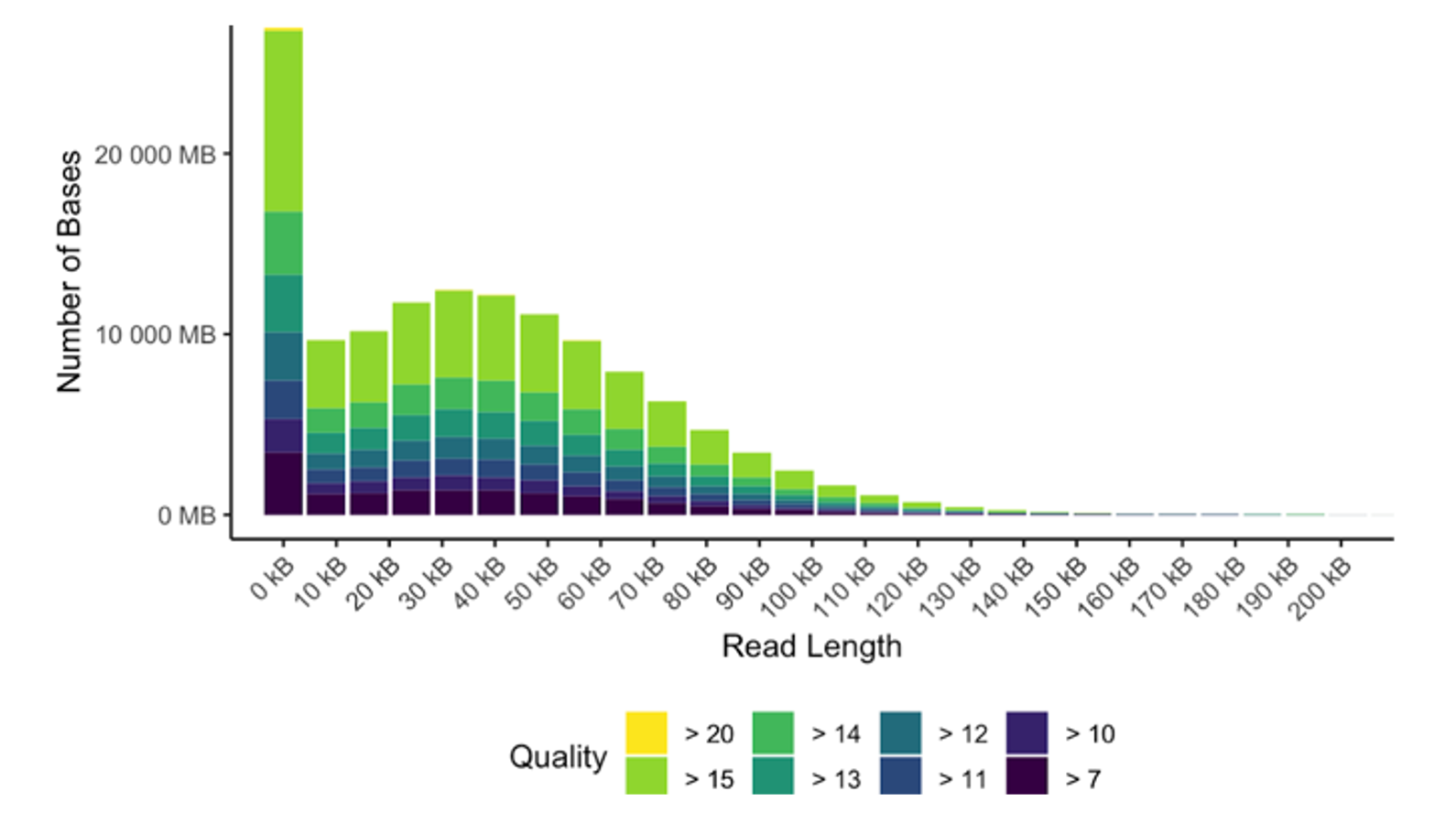
Nanopore sequencing is a third-generation sequencing technology that allows for analysis of DNA and RNA sequences without amplification or modification. As nucleic acids pass through protein nanopores, changes in electric current are detected and subsequently decoded into bases, preserving methylation and modifications to the original sequences. With the PromethION instrument, we generate high-quality datasets with average read lengths of 10–60 kb — suitable for long-read applications such as de novo assembly, structural variant discovery, splice variant discovery, and methylation analysis. Our current workflow is optimized for human samples, and we can make adjustments for select nonhuman sample projects.
We offer cell culture services for projects requiring fibroblast or lymphoblastoid cell lines. We can culture cells from patient-derived samples, ensuring high-quality growth and expansion for downstream genetic analysis or sequencing. Our team can provide guidance on cell line creation and maintenance, helping ensure that your project progresses smoothly.
DNA and RNA extraction and purification services are available on material inputs including cell pellets, saliva, buccal swabs, and blood. See Sample Preparation below for more detail. Contact us to discuss tissue extractions.
For all samples, DNA will be quantified using the Qubit; NanoDrop ratios (260:230 and 260:280) will be used to assess DNA purity and possible extraction carryover contamination; and the Agilent Femto Pulse system is used to evaluate DNA for quality and fragment length. If necessary based on QC results, sample cleanup, shearing, and/or size selection may be performed at additional cost.
Library preparation methods depend on the type of samples being sequenced. We typically use a ligation library prep for DNA sequencing (SQK-LSK114) and a cDNA prep for RNA sequencing (SQK-PCS114). We also offer direct RNA (SQK-RNA004) and rapid library preps (SQK-RAD114).
Additionally, we accept already-prepared libraries for sequencing, but because we are unable to perform our typical QC on those, we cannot guarantee their performance. Results can and do vary based on the quality of the extraction and library preparation methods used. For example, DNA sequencing of samples with protein or RNA carryover may not sequence well.
Using either a rapid (SQK-RBK114.24) or ligation (SQK-NBD114.24) library preparation, we can barcode, pool, and sequence up to 24 samples on a PromethION flow cell. Keep in mind that increasing the number of samples per flow cell will decrease the coverage of each sample. A minimum of 4 samples is required for barcoded runs to ensure optimal data output.
Sequenced libraries may produce datasets of more than 120 Gb, or ~30–40x coverage of the human genome. The raw signal output for all samples will be basecalled using Dorado. We offer several sequencing strategies on the PromethION platform, including:
One hour of analysis is charged per flow cell. This includes basecalling and running samples through our standard in-house analysis pipeline, which calls 5mCG/5hmCG modifications when sequencing native DNA and multiplexed, barcoded samples. Methylations calls are provided as tags in the unaligned bam files that are returned when sequencing is complete. Our standard pipeline is optimized for human samples; data for other organisms may be delivered in other formats and may incur extra analysis charges.
For an added fee, we also offer a range of additional analysis services that can be tailored to meet the specific needs of your study. For example, our team can assist with tasks such as calling additional methylation models, filtering and prioritizing structural variants, identifying repeat expansions, analyzing other complex variants, and data interpretation support for researchers looking to analyze their own data.
We can help you determine which sequencing and analysis services are right for your project. Contact us to set up a consultation and to receive an estimate.
| Service | Internal UW* | External Academic or Nonprofit | Industry |
|---|---|---|---|
| Cell Culture | $116.72 | $134.93 | $148.42 |
| Nucleotide Extraction | $119.62 | $138.29 | $152.11 |
| Sample QC | $55.79 | $64.49 | $70.94 |
| Sample Cleanup | $41.99 | $48.54 | $53.40 |
| Shearing | $48.85 | $56.47 | $62.11 |
| Size Selection | $99.38 | $114.88 | $162.37 |
| Library Preparation | $259.72 | $300.24 | $330.27 |
| Flow Cell | $1,238.95 | $1,432.22 | $1,575.45 |
| Barcoding – Rapid Prep (per sample) | $41.77 | $48.29 | $53.12 |
| Barcoding – Ligation Prep (per sample) | $84.16 | $97.28 | $107.01 |
| Barcoding – Pool (4–24 samples per pool) | $106.24 | $122.81 | $135.09 |
| Analysis (per hour) | $218.36 | $252.42 | $277.66 |
*Staff and faculty at the University of Washington paying with a UW Worktag receive the Internal rate; all others are charged the External or Industry rate. Rates are subject to change without notice.
Genome coverage and read length distributions in your aligned data are highly dependent on the quality of input material. The following guidelines will help ensure you receive the highest quality data possible.
We currently accept several sample input types, including DNA, RNA, blood, saliva, buccal swabs, and cell pellets. For other sample types, please inquire directly. Follow the guidelines below for your specific sample type.
Run a standard or pulsed-field gel, BioAnalyzer, or TapeStation to visualize DNA quality and length before submitting. If your sample looks degraded, re-extract the sample or plan for shorter read lengths. While ~30–40x coverage of the human genome may be achieved with one PromethION flow cell, poor quality DNA will not yield these results. If submitting old, highly fragmented, or otherwise degraded DNA, you should expect ~10–20x coverage.
Important notes:
For direct RNA submission, contact us to discuss the guidelines.
Our recommendation for stabilizing blood samples is to use a purple top EDTA blood collection tube. A minimum of 1 mL of blood is required for high molecular weight DNA extraction.
Saliva and buccal samples can be shipped at room temperature. We typically see higher DNA yields from saliva versus buccal samples. Our current protocol uses the following DNA Genotek collection kits for research:
To limit contamination when collecting saliva, it's important that the individual does not eat or drink for 30 minutes prior to collection. Our current extraction method for saliva samples is the automated QIAsymphony DSP DNA Midi Kit.
Submit between 1 x 10e5 and 1 x 10e7 cells per pellet. Cells must be frozen as pellets in Eppendorf tubes. Microcentrifuge tubes should be double-packed into larger conical tubes and shipped in a polystyrene box filled at least halfway with dry ice. Send these samples using next-day delivery service according to the shipping instructions shown below.
If you would like to drop off your samples in person, we accept delivery 10 am – 4 pm on weekdays.
For shipping, plan for weekday delivery only, 10:00 am – 4:00 pm.* All samples should be delivered to:
University of Washington Nanopore Sequencing Core
c/o Joy Goffena
1705 NE Pacific St.
HSB RM H-458
Seattle, WA 98195
For all deliveries:
* Do not send packages for first morning delivery. FedEx and UPS do not have early morning access to our building, so deliveries arriving prior to 9 am, including FedEx First Overnight deliveries, may be delayed by a day or more.
* Do not send packages to arrive on weekends or after 4:00 pm on weekdays.
* Do not ship samples without contacting us first.