University of Washington | Department of Pediatrics | Department of Laboratory Medicine & Pathology
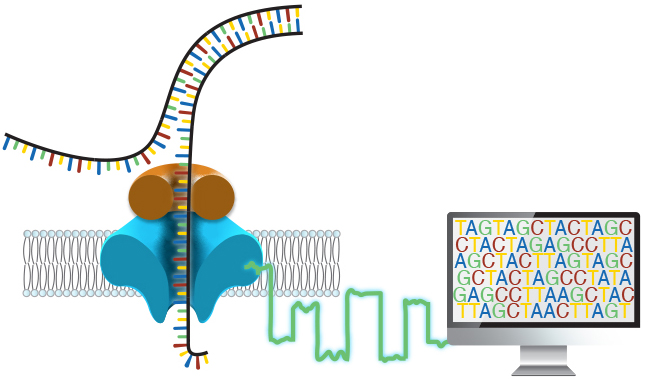
Around 50% of individuals with a suspected genetic disorder remain undiagnosed after a complete clinical evaluation, which often takes years to complete. We believe this burden on patients and families is simply too high. In the Miller Lab, our goals are threefold: to improve the efficiency and effectiveness of genetic testing, to expand access to genetic testing, and to better understand human genetic disease through the identification and characterization of novel disease-causing variation.